A mouth bacteria has starring role in colorectal cancer, study finds Premium
The Hindu
Researchers have identified a subtype of Fusobacterium bacteria to be linked to colorectal cancer, potentially paving the way for early detection and targeted treatments.
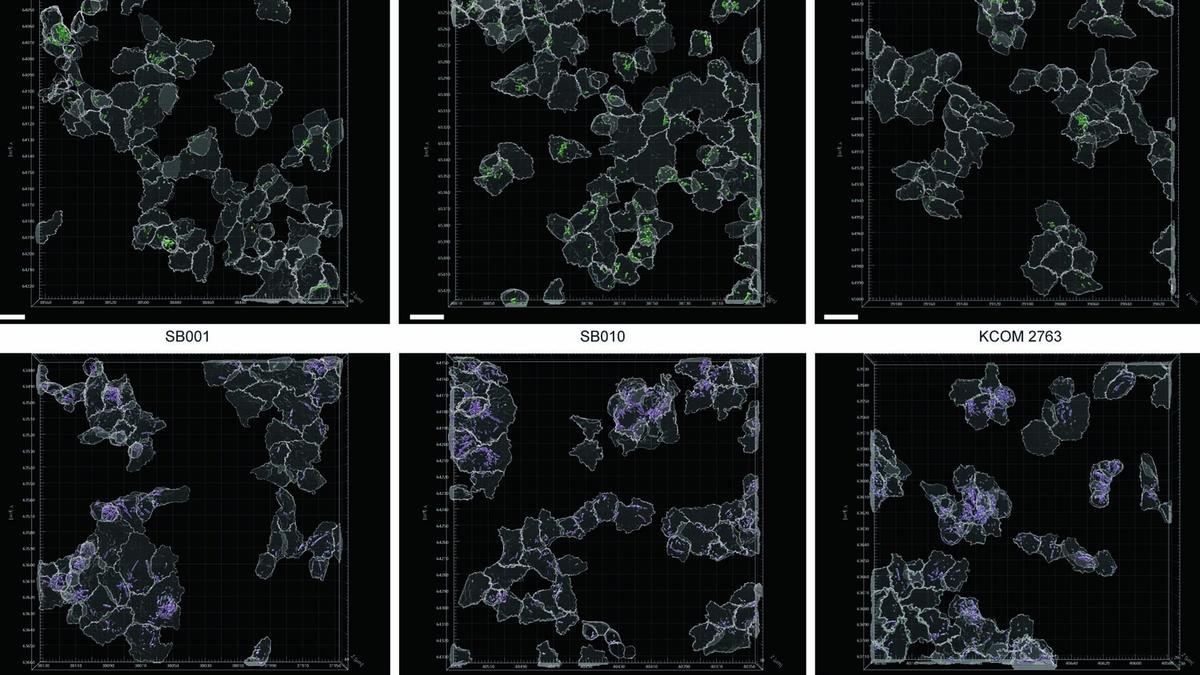
The bacteria known as Fusobacterium nucleatum live in the human mouth and are rarely found elsewhere. But in cases of cancer of the colon or the rectum, the bacteria are found in tumours in the gut, where they help cancer cells escape from the immune system and spread to other parts of the body.
In a new study, a group of researchers from the Fred Hutchinson Cancer Center in the U.S. has identified a distinct subtype of the bacterium that’s found in relatively greater quantities in colorectal cancer (CRC) tumours.
CRC is the seventh most common type of cancer in India, where the number of cases rose by 20% from 2004 to 2014. Worldwide, the overall CRC incidence has declined but, experts wrote in the journal Science last year, the incidence of age-adjusted early-onset CRC “has risen at an alarming rate of 2-4% in many countries, with even sharper increases in individuals younger than 30 years.”
According to the team’s experiments, described in a paper in Naturein March, some genetic factors could be boosting Fusobacterium’s ability to associate with cancers of the gut. The team also showed that when mice were infected with this type of Fusobacterium, their intestines developed precancerous formations called adenomas.
Experts said the study’s findings could be used in future to develop tests to detect CRC early and develop targeted treatment options.
The researchers began by culturing Fusobacterium bacteria collected from 130 human CRC tumours in the laboratory. Then they mapped the entire genetic composition of the isolated bacteria and found that out of the four known Fusobacterium nucleatum subspecies, only Fusobacterium nucleatum animalis (Fna) was significantly associated with CRC tumours.
Individual members of the same species have slightly different DNA. Pangenomic analysis helps researchers map all the genes in a species as well as those parts of the genome that some but not all members of the species have. This part is called the accessory genome. The members of a species can be further subclassified depending on the accessory genomes they have.

Gaganyaan-G1, the first of three un-crewed test missions that will lead up to India’s maiden human spaceflight, is designed to mimic - end to end - the actual flight and validate critical technologies and capabilities including the Human-rated Launch Vehicle Mark-3 (HLVM3), S. Unnikrishnan Nair, Director, Vikram Sarabhai Space Centre (VSSC), has said










